주제 4 고해상도 혈관벽 자기공명영상 병변 검출

Scientific overview (연구 배경 및 중요성)
Vessel wall magnetic resonance imaging (VW-MRI) enables direct visualization of the vessel wall and its pathology with high spatial resolution and excellent soft tissue contrast. It has been increasingly used as an adjunct to digital subtraction angiography and MR angiography in establishing the etiology for intracranial stenosis. Estimating plaque burden and vulnerability in patients with intracranial atherosclerotic disease is essential in risk assessment and guiding clinical management. In addition, VW-MRI may reveal the characteristic features of intimal flap, double lumen, or intramural hematoma seen in dissection. While lesion segmentation on VW-MRI is important in lesion evaluation and monitoring disease progression, segmentation of mural lesions on VW-MRI is limited by the clinically feasible spatial resolution of VW-MRI and complex geometry of vascular pathology. Automatic lesion segmentation on VW-MRI will improve reproducibility and enable subsequent analysis in the quantitative measurement and radiomics pipeline.
Challenge questions (문제 정의)
Develop an AI model to accurately segment vessel wall lesion on high resolution T1-weighted image
Data description (데이터 설명 – 데이터 셋 구성, 형식, 특징)
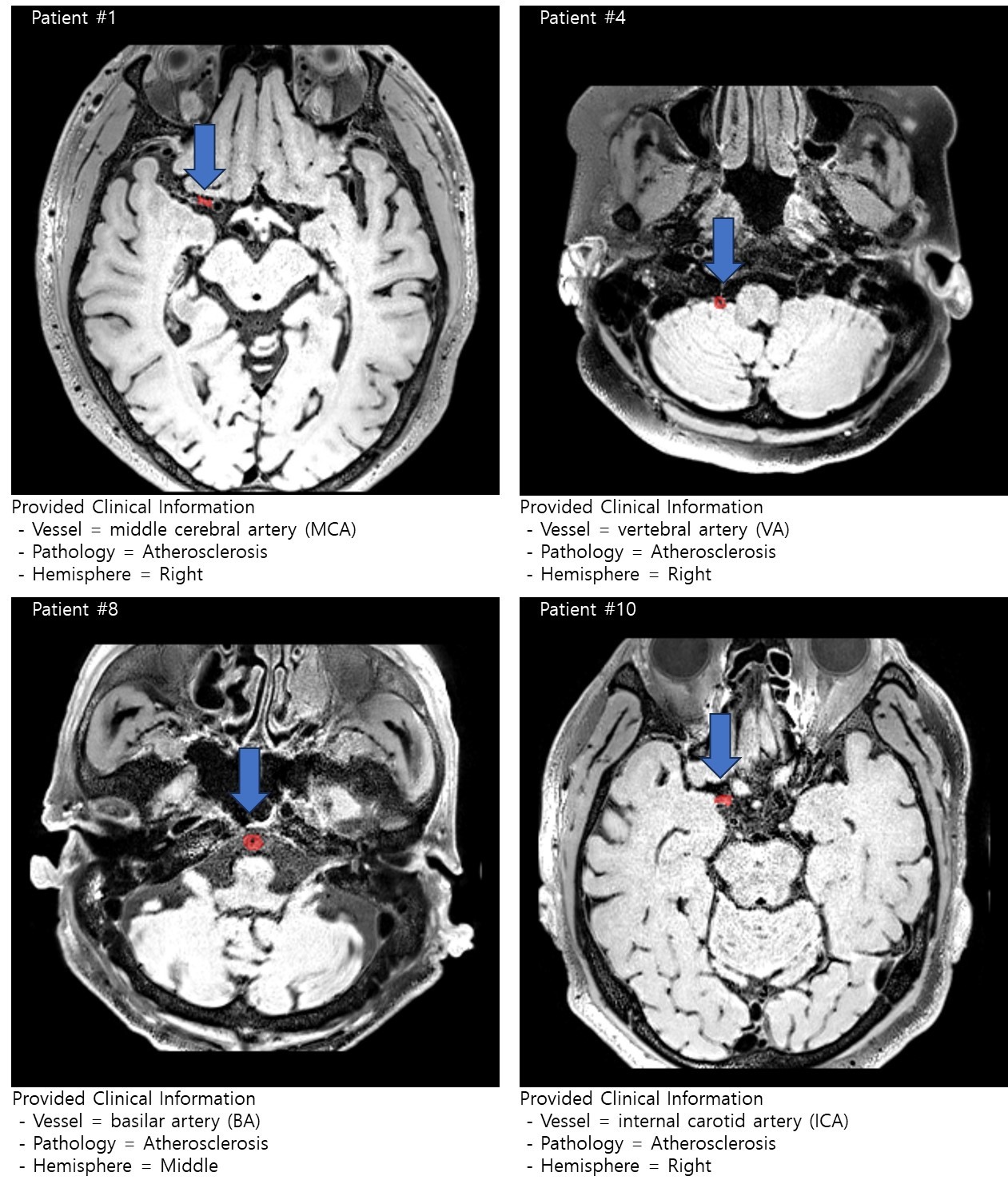
- (1) Data Set : 80 of stroke patients (50 and 30 for Training and Validation, respectively)
-
(2) Clinical Data and Label : Location of the lesion in vessel category and pathology of the lesion
- Vessel category: 1 = ICA, 3 = MCA, 5 = BA, 6 = VA
(ICA = internal carotid artery, MCA = middle cerebral artery, BA = basilar artery, VA = vertebral artery)
- Pathology: 1 = Atherosclerosis, 4 = Vasculitis
- Hemisphere: Left / Middle / Right indicate the hemisphere of lesion location - (3) Image Data : T1-weighted image and Lesion mask (lesion mask is target label)
The T1-weighted image was acquired using 3.0-T MRI scanner (Skyra, Siemens; TR/TE = 900ms/15ms, flip angle = 120, Acquisition Matrix = 320 x 320, FOV = 192 x 192, slice thickness = 0.6mm without gap).
Infarct regions were manually segmented on the T1-weighted image by an expert neuroradiologist.
The brain MRI and lesion mask (region-of-interest, ROI) are provided in compressed NifTi format (.nii.gz). Surface (e.g., Face) of image data was eliminated for anonymization. All the image data will be provided its acquisition space without preprocessing.
 [T1_Space] Folder
[T1_Space] Folder
- (1) T1_FOV.nii.gz
- (2) ROI.nii.gz (training data will be provided, i.e., VW_Subject001 ~ VW_Subject050)/li>
Evaluation matrix (정량적 평가 방법)
(1) Requirements for participants in submission (excel and NifTi format files)
- Submit excel (list of filename) and NifTi files (predicted lesion mask by model, binary)
- NifTi files (i.e., predicted lesion mask) should be contained in a file (e.g., OOO.zip / OOO.tar.gz) (e.g., VW_Subject001_ROI.nii.gz ~ VW_Subject080_ROI.nii.gz)
(2) Evaluation
- Average dice similarity coefficient (DSC) in validation data




